
This research project is conducted in a close collaboration with Prof. Vladimir Privman
PI: Evgeny Katz, Co-PI: Vladimir Privman
Enzyme-Based
Biomolecular Computing

Molecular and biomolecular
logic gates
and their networks processing chemical input signals similarly to
computers
received high attention and were rapidly developed in the last decade.
Being a
subarea of unconventional computing, they can process chemical
information
mimicking Boolean logic operations using binary definitions (1,0;
YES/NO) for concentrations of reacting
species. Using this approach,
chemical reactions could be reformulated as information processing
steps with
built-in logic operations. Then, the chemical processes could be
programmed
similar to computer programming yielding networks performing several
logic
operations.
As a network that processes
information becomes large, and the information is processed in greater
quantities and at higher complexity, noise inevitably builds up and can
ultimately degrade the useful “signal” which is the intended result of
the
computation. One then has to develop approaches to achieve what is
known as
“fault-tolerant” information processing that involves noise control and
suppression. One notable example is the “robustness” of many complex
processes
in cell functions.
Presently, we are aware of
three
primary fault-tolerant information processing paradigms. The first is
the
analog/digital electronics paradigm of the Si-chip technology in modern
computers. We know how to design such devices and they have been
successfully
built. Living organisms are the second paradigm: While we obviously
know that
this paradigm leads to scalability, we do not yet fully understand it,
though
significant strides have been made in Systems Biology to explore the
structure
and functioning of biological “networks.” The third, recent paradigm,
involves
massive parallelism: quantum (quantum computing) or ensemble (variants
of DNA
computing), both in the preliminary research stages.
Biochemical
computing — in
our case based on enzymatic reactions1-3 — attempts
to
process information with biomolecules and biological objects.4-13
However, the information processing paradigm assumed, has, in most
cases, been
that of the ordinary electronics. Indeed, most biochemical computing
studies
attempt to realize and, most recently, network “gates” that mimic
Boolean
digital logic.14,15
Networks with computational
steps that
solely involve biochemical processes,16,17 are being
researched for
new technological capabilities: multi-input biosensors with new
functionalities,18-20 as well as approaches that allow
removing the
batteries from, and generally reducing the need for inorganic leads and
electrical power supply for those stages of information processing that
occur
during biomedical testing, implantable devices, and other fast decision
making
steps in applications. Futuristic ideas for applications of biochemical
logic
include more direct brain-computer and body-computer interfacing for
both
reading out and inputting information, and generally erasing the
barrier
between the inorganic and organic information processing in computer
device
functioning.
The
primary present challenge in
the field of biochemical computing can be summarized as follows. Recent
studies
took the field somewhat beyond the earlier set of works that have
realized
simple gates mimicking two-input, one-output Boolean functions.
Specifically,
few-gate networking has been accomplished,16,17 first steps
in the
analysis of network scalability have been reported very recently,21
and first attempts at “smart” interfacing with ordinary electronics
have been
initiated.22-25 In these studies, biocomputing based on
enzymatic
reactions has emerged as an appealing approach for information
processing due
to the specificity and other useful chemical-kinetics properties of
enzyme
reactions.
However, these advances
have also set
the stage for new challenges. In particular, it has been realized that
large-scale networking and fault-tolerance cannot be achieved without
the
development of a “toolbox” of new network elements, specifically,
filters,
signal splitters (copying), signal balancers, resetting functions, etc.
These
non-Boolean network elements for biochemical computing might not follow
too
closely the analogy with ordinary electronic devices. In fact, concepts
borrowed from Nature, including the delayed identity — part of the
feed-forward network motif,26 or “memory” properties27
have recently received attention in information processing network
studies. The
present research program attempts to address these challenges, building
on the
earlier works in the fields of chemical and biochemical computing.
At present there are
chemical-computing studies that have used molecular or supra-molecular
systems28‑44
to mimic processes typical for electronic computing devices such as
simple
Boolean logic operations AND, OR, XOR, etc.,45-63 as well as more complex
systems
including molecular comparator, digital demultiplexer,
encoder-decoder, keypad lock,
write-read-erase memory units, half-adder/half-subtractor or
full-adder/full-subtractor,64-86 etc.
Chemical systems are, in principle, capable of performing computations
at the
level of a single molecule87 resulting in nano-scaling of
the
computing units88 and allowing parallel computation
performed by
molecules involved in various chemical reactions.89 However, one of the most important
challenges in the
chemical computing is networking and fault-tolerance.1,15
Complex
multi-component chemical logic systems usually require ingenious
molecular
assemblies to allow compatibility between information processing
sub-units28,77
and perform rather simple functions despite their extreme synthetic
complexity.90
Biomolecular systems, on
the other
hand, offer the advantage of specificity (of being very selective) in
their
chemical functions and therefore being more usable in complex “chemical
soup”
environments. As a result, complex enzyme-based information processing
units
could be more easily scaled up giving rise to artificial biocomputing
networks
performing various logic functions and mimicking natural biochemical
pathways.16,17,21,42,91
An added advantage of biomolecular computing systems is the capability
to
process biochemical information received in the form of chemical
signals directly from biological
systems and the ability to operate in a biological environment.92
This is important for interfacing of the resulting biochemical-logic
“devices”
with processes in living organisms,93 for potential
biomedical
applications.
Recent experimental
advances in
enzyme-based biocomputing have included not just experimental
demonstrations of
several single Boolean gates, such as AND,
OR, XOR, Inhibit,
etc.,1-4
but also networking of several, up to 3-5 presently, gates.14,16,17,21,24
Similar logic operations were also realized using non-biological
chemical
systems.64-85 However, the biochemical systems offered
relative
simplicity of the assembled logic schemes. Still, ultimately the
increasing
complexity of enzyme-based logic networks will require exploration of
noise
suppression approaches in biochemical logic-gates networks. Fault
tolerance
within the analog/digital information processing paradigm is
accomplished by
gate optimization for suppression of the “analog” noise amplification1,94
as well as by network design and/or network topology.21 For
larger
networks, another, “digital” mechanism95 of noise
amplification
emerges. It is combated by redundancy in network design and requires
truly
digital information processing with appropriate network elements for
filtering,
signal splitting, etc.
The present sizes of the
biochemical
computing networks have already allowed exploration of aspects of
design and
optimization issues related to suppression of the “analog” noise
amplification.
The work on this topic was initiated in our recent publications.1,21,94,95
The field is presently wide open for research developments. The
following brief
description of our
recent results attempts to summarize the group activity in the project.
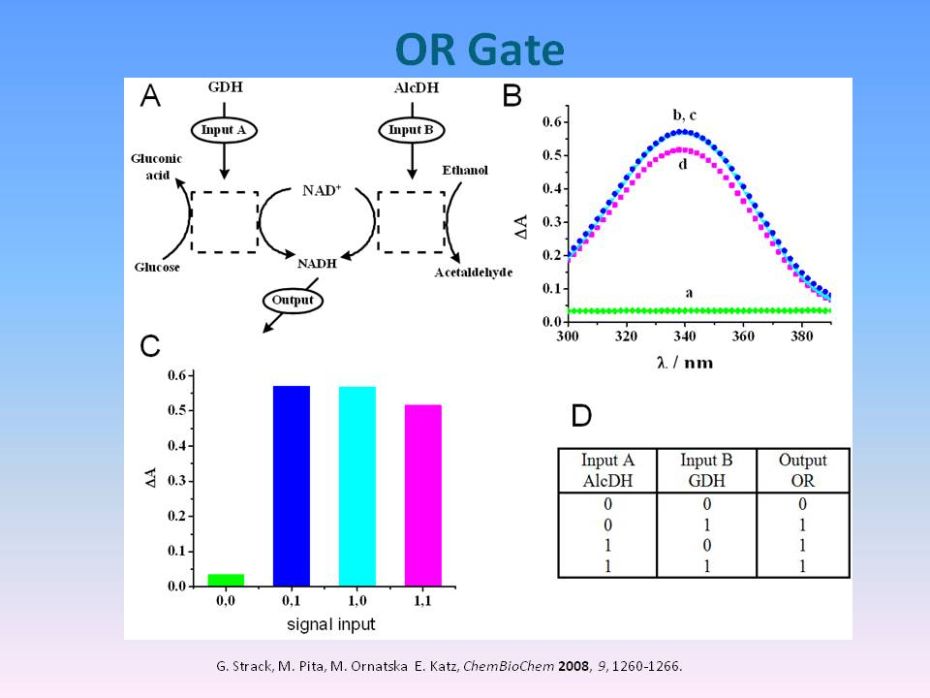
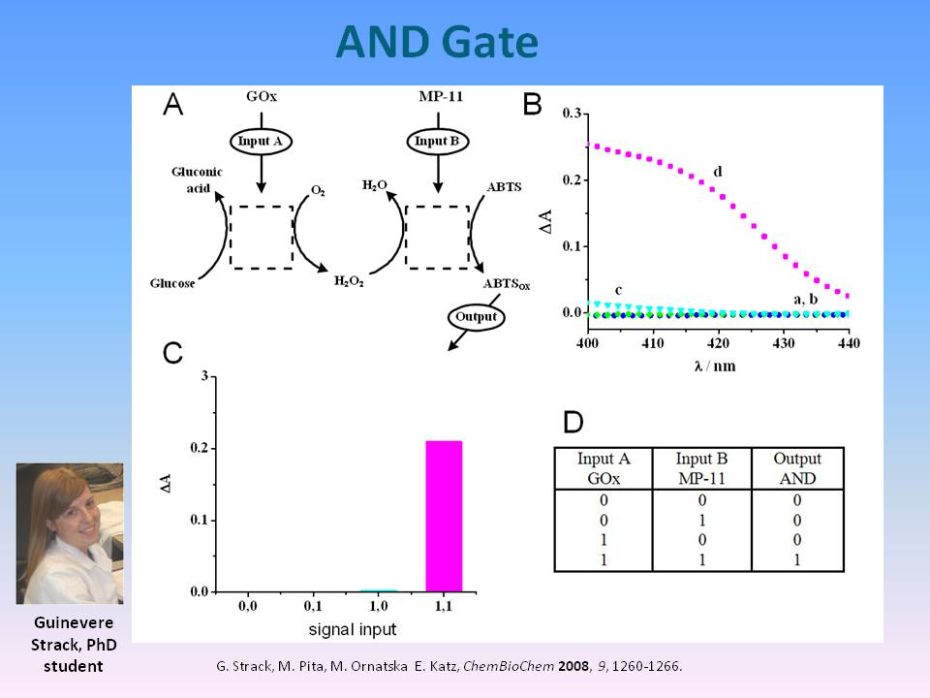 |
Boolean Logic Gates
Using Enzymes
as Input Signals
G.
Strack, M. Pita, M. Ornatska,E. Katz, Boolean logic gates using enzymes
as input signals. ChemBioChem,
2008, 9, 1260-1266. |
 |
 |
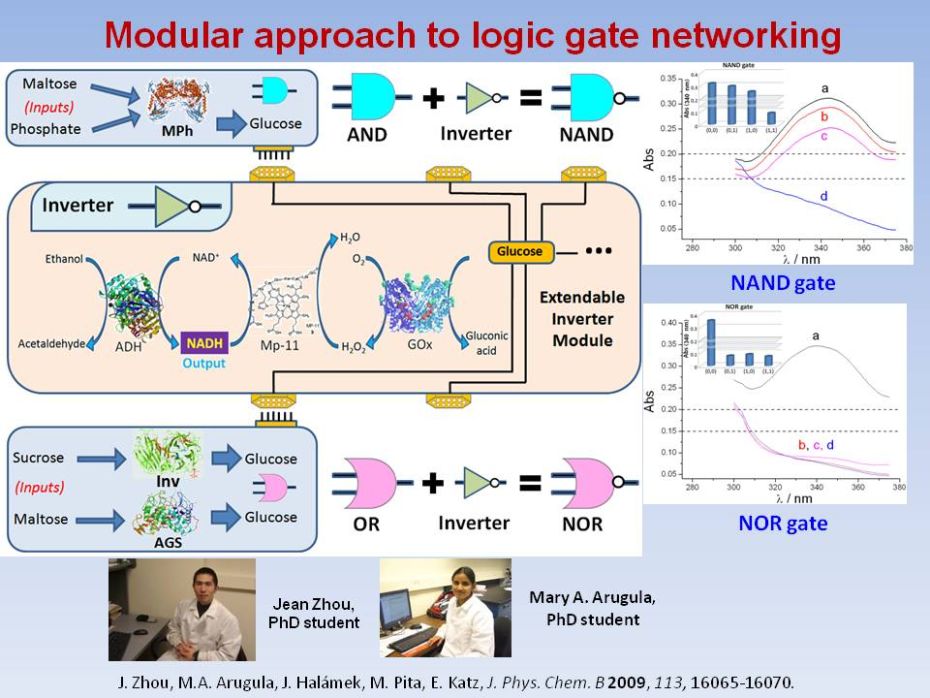 |
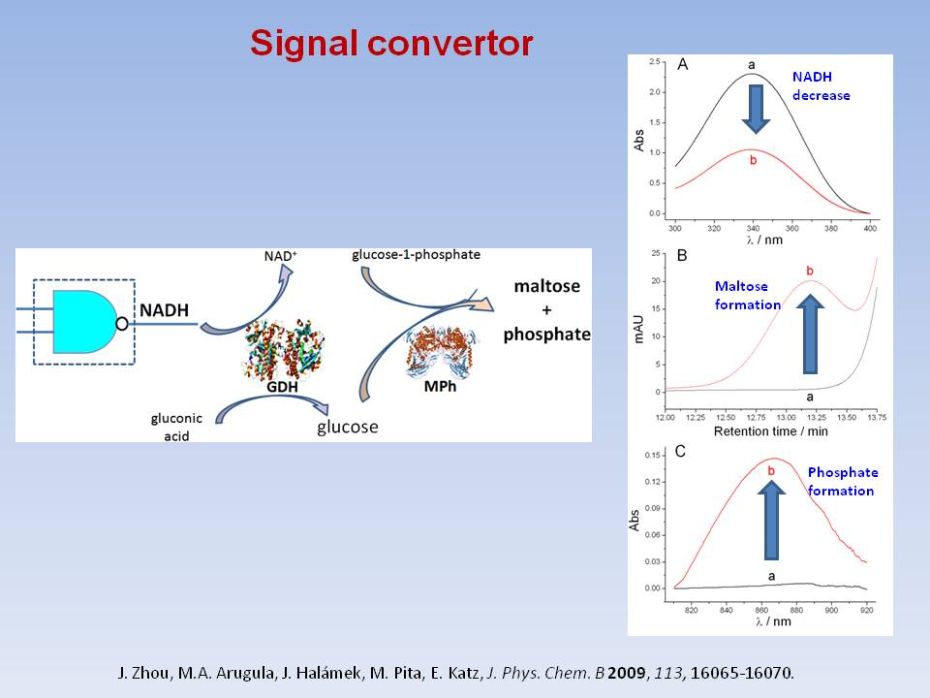
Enzyme-based
NAND and NOR logic gates with modular design
The
logic gates NAND / NOR were mimicked by enzyme
biocatalyzed reactions activated by sucrose, maltose and phosphate. The
sub-units performing AND / OR Boolean logic operations were
designed using maltose phosphorylase and cooperative work of
invertase/amyloglucosidase, respectively. Glucose produced as the
output signal from the AND / OR sub-units was applied as the
input signal for the INVERTER
gate composed of alcohol dehydrogenase, glucose oxidase,
microperoxidase-11, ethanol and NAD+, which generated the
final output in the form of NADH inverting the logic signal from 0 to 1 or from 1 to 0. The final output signal was
amplified by a self-promoting biocatalytic system. In order to fulfill
the Boolean properties of associativity and commutativity in logic
networks, the final NADH output signal was converted to the initial
signals of maltose and phosphate, thus allowing assembling of the same
standard units in concatenated sequences. The designed modular
approach, signal amplification and conversion processes open the way
towards complex logic networks composed of standard elements resembling
electronic integrated circuitries.
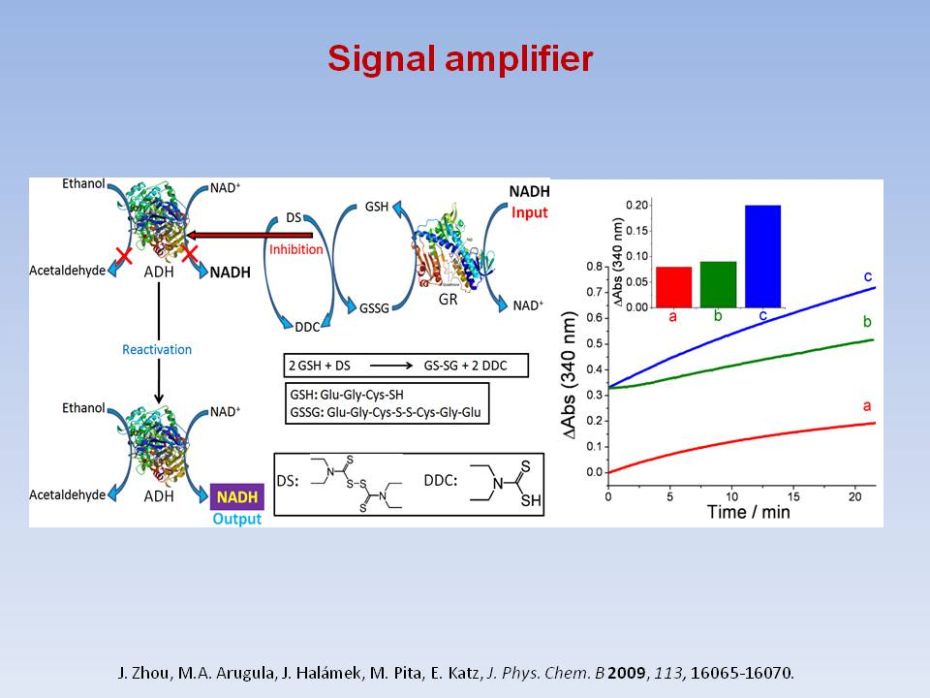
J. Zhou, M.A. Arugula, J. Halámek, M. Pita, E. Katz, Enzyme-based NAND and NOR logic gates with modular design. J. Phys. Chem. B 2009, 113, 16065-16070. |
 |
 |
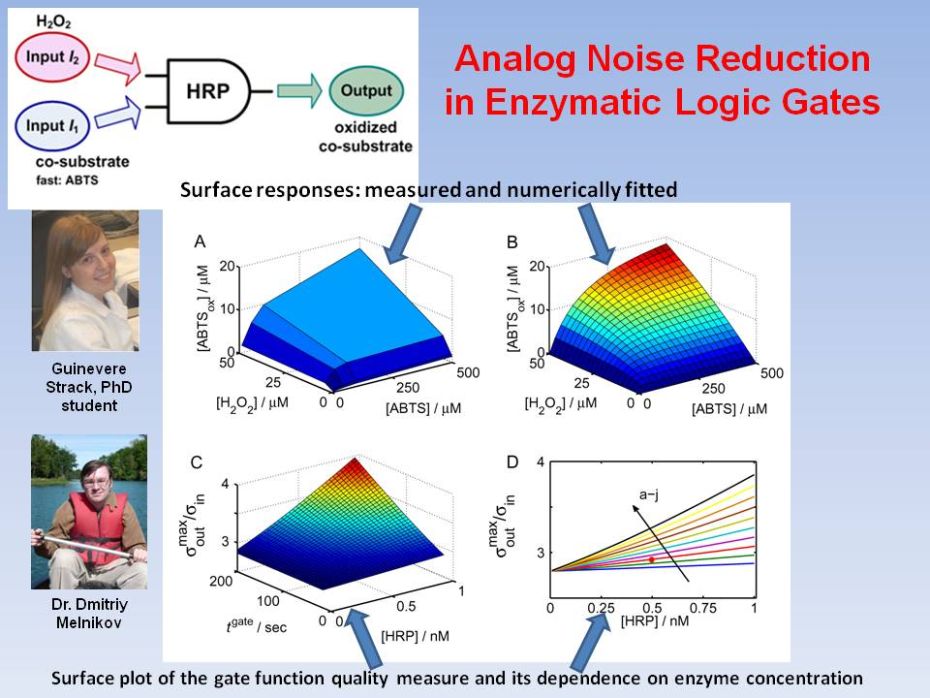 |
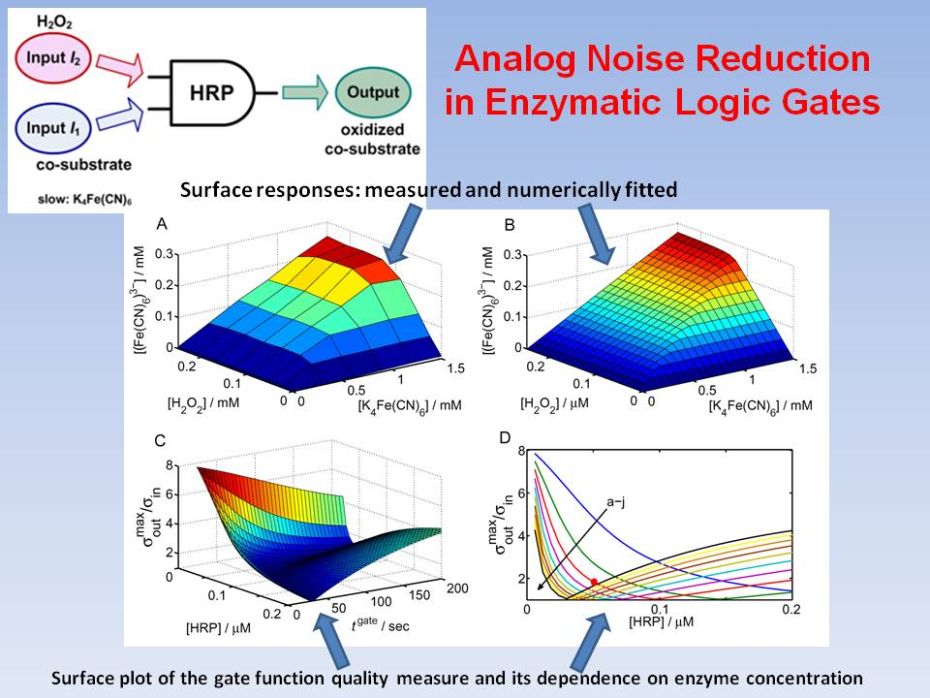
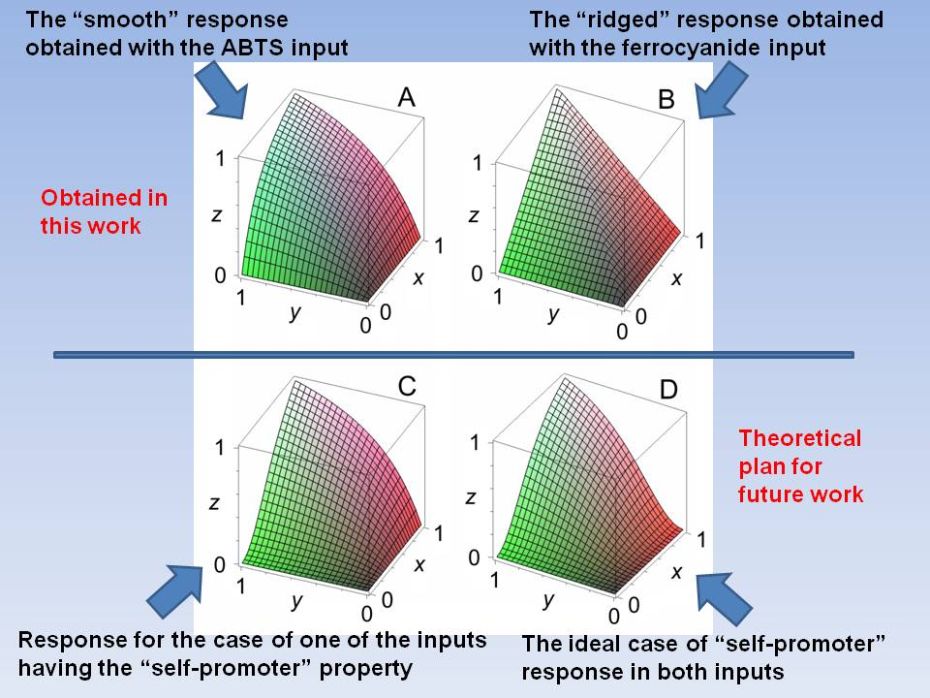
Analog Noise
Reduction in Enzymatic
Logic Gates
In this work we demonstrate both experimentally and theoretically that the analog noise generation by a single enzymatic logic gate can be dramatically reduced to yield gate operation with virtually no input noise amplification. We demonstrate that when a co-substrate with a much smaller affinity than the primary substrate is used, a negligible increase in the noise output from the logic gate is obtained as compared to the input noise level. Our general theoretical conclusions were confirmed by experimental realizations of the AND logic gate based on the enzyme horseradish peroxidase using hydrogen peroxide as the substrate, with 2,2'‑azino‑bis(3-ethylbenzthiazoline-6-sulphonic acid) (ABTS) or ferrocyanide as co‑substrates with vastly different rate constants. D. Melnikov, Strack, M. Pita, V. Privman, E. Katz, Analog noise reduction in enzymatic logic gates. J. Phys. Chem. B 2009, 113, 10472-10479. |
 |
 |
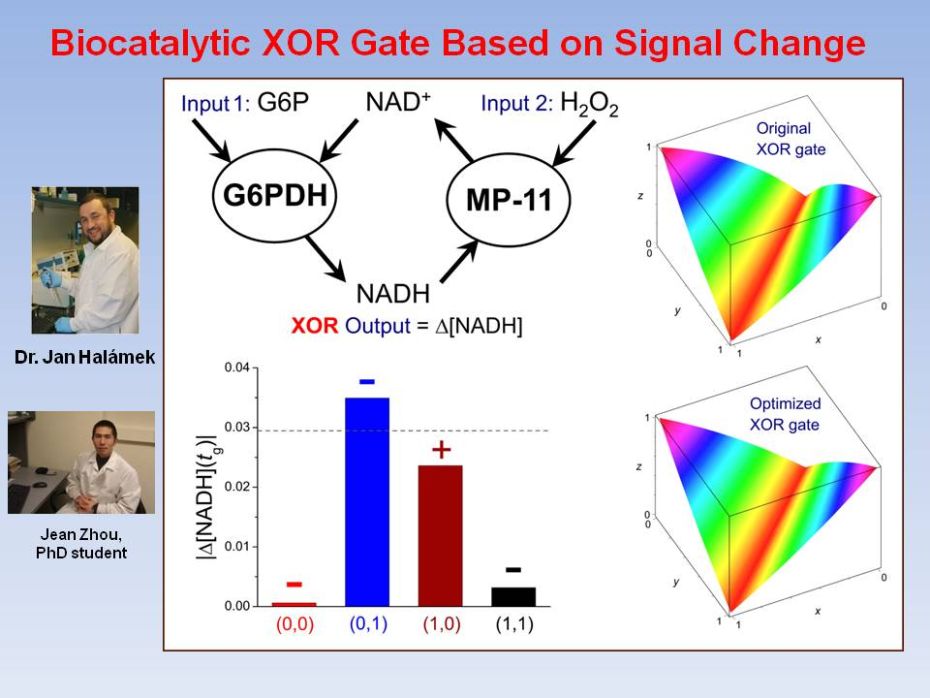 |
Realization
and Properties of Biochemical-Computing Biocatalytic XOR Gate Based on
Signal
Change We consider a realization of the XOR logic gate in a system involving
two competing biocatalytic reactions, for which the logic-1 output is defined by these two
processes causing a change in the optically detected signal. A model is
developed for describing such systems in an approach suitable for
evaluation of the analog noise amplification properties of the gate and
optimization of its functioning. The initial data are fitted for gate
quality evaluation within the developed model, and then modifications
are proposed and experimentally realized for improving the gate
functioning.
V. Privman, J. Zhou, J. Halámek, E.
Katz, Realization and properties of biochemical-computing biocatalytic
XOR gate based on signal change. J.
Phys. Chem. B 2010, 114, 13601-13608.
|
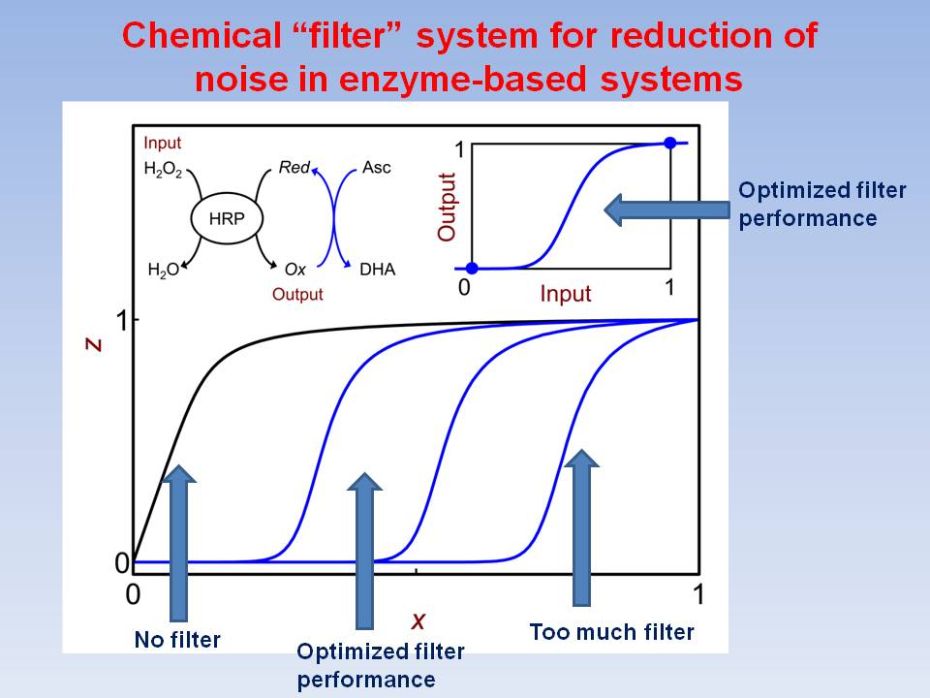 |
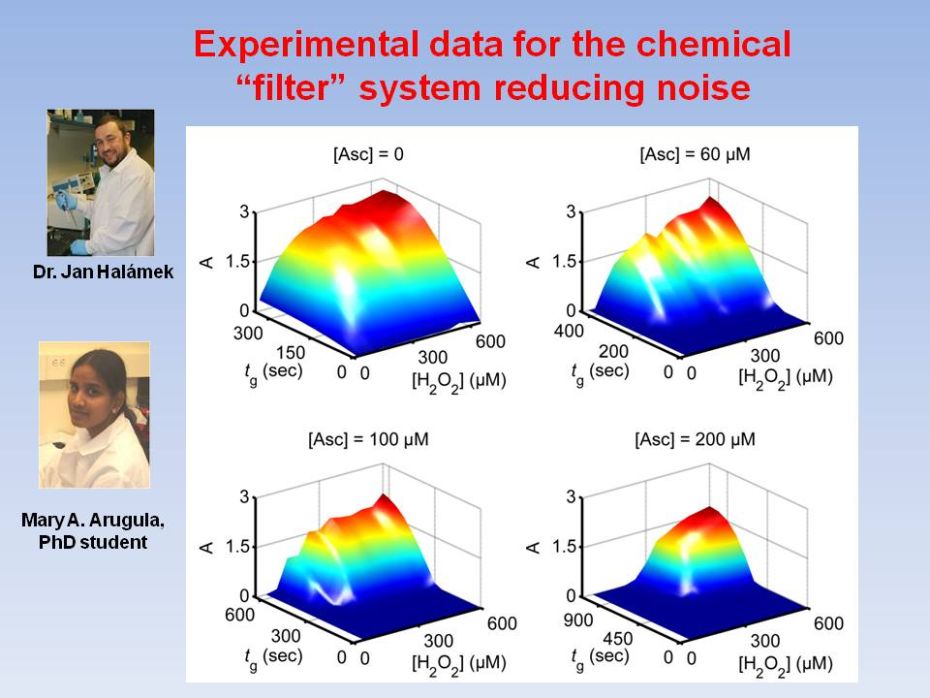
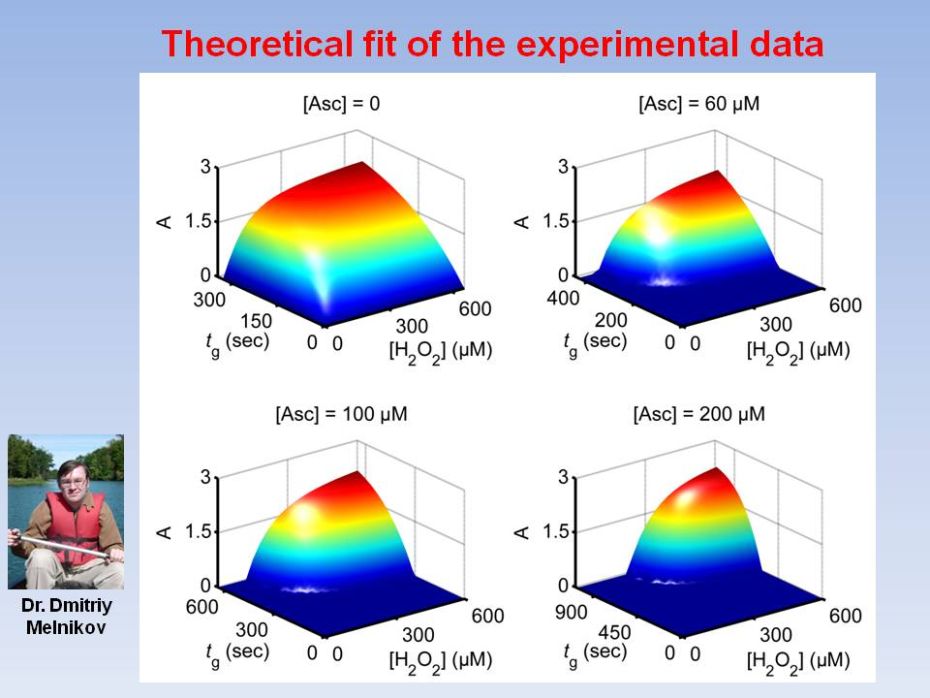
Biochemical
Filter with Sigmoidal Response:
The first
realization of a designed, rather than natural, biochemical filter
process is
reported and analyzed as a promising network component for increasing
the
complexity of biomolecular logic systems. Key challenge in biochemical
logic
research has been achieving scalability for complex network designs.
Various
logic gates have been realized, but a "toolbox" of analog elements
for interconnectivity and signal processing has remained elusive.
Filters are
important as network elements that allow control of noise in signal
transmission and conversion. We report a versatile biochemical
filtering
mechanism designed to have sigmoidal response in combination with
signal-conversion process. Horseradish peroxidase-catalyzed oxidation
of chromogenic
electron donor by H2O2, was altered by adding ascorbate,
allowing to selectively suppress the output signal, modifying the
response from
convex to sigmoidal. A kinetic model was developed for evaluation of
the
quality of filtering. The results offer improved capabilities for
design of
scalable biomolecular information processing systems.
V. Privman, J. Halámek, M.A. Arugula, D. Melnikov, V. Bocharova, E. Katz, Biochemical filter with sigmoidal response: Increasing the complexity of biomolecular logic. J. Phys. Chem. B 2010, 114, 14103-14109.
|
 |
 |
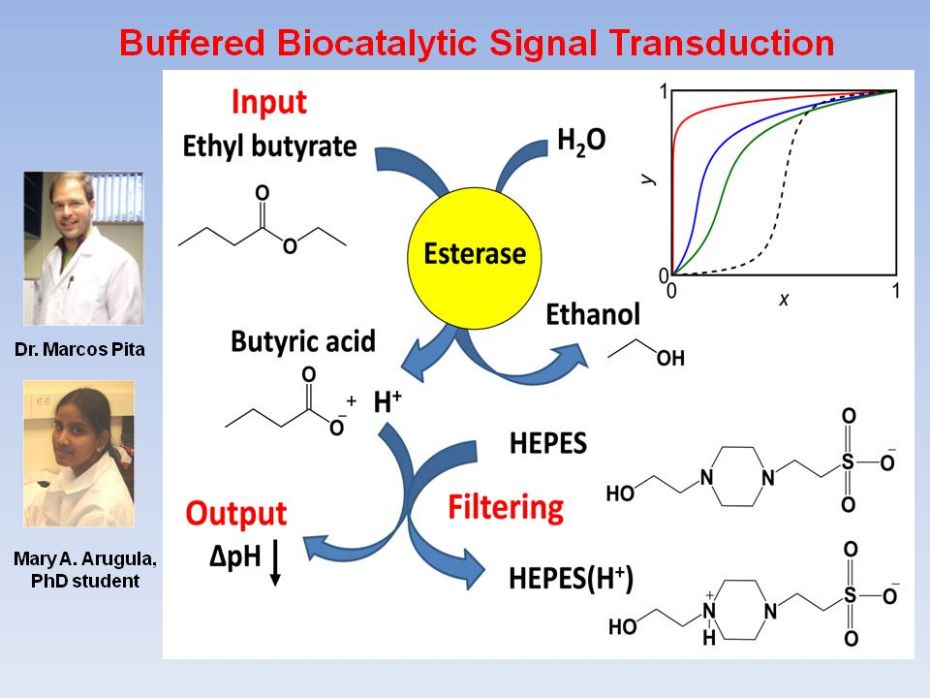 |
Towards
Biochemical Filter with Sigmoidal Response to pH Changes
We
realize a biochemical filtering process
based on the
introduction of a buffer in a biocatalytic signal-transduction logic
system
based on the function of an enzyme, esterase. The input, ethyl
butyrate, is
converted into butyric acid—the output signal, which in turn is
measured by the
drop in the pH value. The developed approach offers a versatile
"network
element" for increasing the complexity of biochemical information
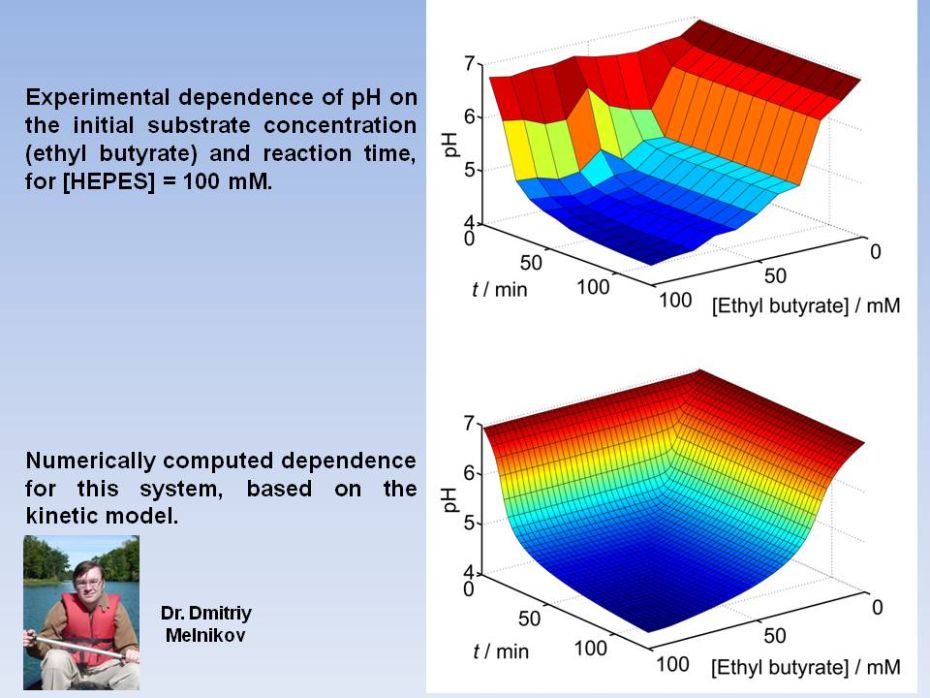
processing systems. Evaluation of an optimal regime for quality
filtering is
accomplished in the framework of a kinetic rate-equation model.
The reaction biocatalyzed by an enzyme,
here esterase,
results in the hydrolysis of ethyl butyrate (the logic Input)
to yield
butyric acid which releases H+ ions upon dissociation. A limited
quantity of a buffer, here HEPES, if introduced, consumes most of the
biocatalytically produced H+ ions when the input is applied at a low
concentration. The pH change (the logic Output,
measured by the pH drop,
as indicated by an arrow) sets in when the biocatalytically produced H+
ions overwhelm the buffer. The biocatalytic process and buffering
combined,
yield a sigmoidal dependence of the pH change as a function of the
input
concentration. The inset illustrates the onset of the sigmoidal
response in our
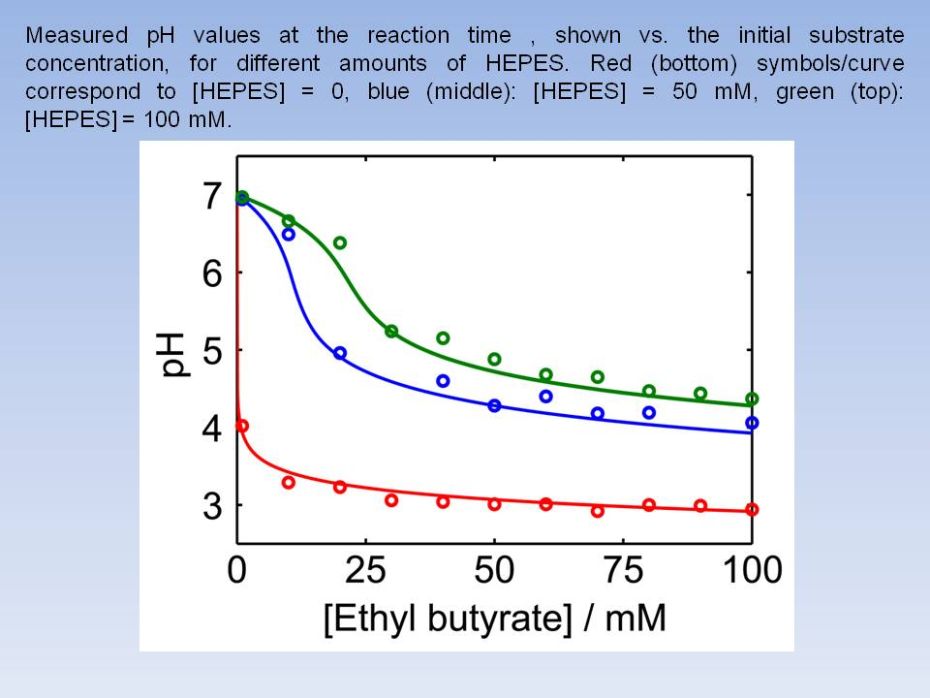
experimental system. The solid curves show the output, y,
vs. the input, x, properly redefined/rescaled to vary in
the "binary-logic
ranges" from 0 to 1, as explained in the text. Experimental data were
fitted by using rate equations appropriate for the processes involved,
and the
results are shown, here for the reaction time 120 min, for
increasing
buffer (HEPES) concentrations. The top (red) curve corresponds to
[HEPES] = 0; middle (blue): [HEPES] = 50 mM,
bottom
(green): [HEPES] = 100 mM. The dashed black curve does
not
correspond to experimental data but rather illustrates a desirable,
"ideal" filter response with small slopes at both binary logic points 0
and 1, and with a steep, symmetrically
positioned inflection
region in the middle. M. Pita, V. Privman, M.A.
Arugula, D. Melnikov, V. Bocharova, E.
Katz, Towards biochemical
filter with sigmoidal response to pH changes: Buffered biocatalytic
signal transduction. PhysChemChemPhys 2011,
in press (DOI:
10.1039/c0cp02524k). |
 |
 |
1.
V. Privman, G. Strack, D. Solenov,
M. Pita,
E. Katz, Optimization of enzymatic biochemical logic for noise
reduction
and scalability: How many biocomputing gates can be interconnected in a
circuit? J. Phys. Chem. B 2008, 112,
11777-11784.
2. G.
Strack, M. Pita, M. Ornatska, E. Katz, Boolean logic gates using
enzymes as
input signals, ChemBioChem 2008, 9, 1260-1266.
3. R.
Baron, O. Lioubashevski, E. Katz, T. Niazov, I. Willner, Two coupled
enzymes
perform in parallel the “AND” and “InhibAND” logic gates operations, Org. Biomol. Chem. 2006, 4,
989-991; R. Baron, O.
Lioubashevski, E. Katz, T. Niazov, I. Willner, Logic gates and
elementary
computing by enzymes, J. Phys. Chem. A
2006, 110, 8548-8553.
4. S. Sivan, N. Lotan, A
biochemical logic
gate using an enzyme and its inhibitor. Part I: The inhibitor as
switching
element, Biotechnol. Prog. 1999, 15, 964-970;
S. Sivan, S. Tuchman, N. Lotan, A biochemical logic gate
using an
enzyme and its inhibitor, Biosystems 2003,
70, 21-33.
5. A.S. Deonarine, S.M. Clark, L.
Konermann,
Implementation of a multifunctional logic gate based on
folding/unfolding
transitions of a protein, Future
Generation Comp. Syst. 2003, 19,
87-97.
6. G. Ashkenazi, D.R. Ripoll, N.
Lotan,
H.A. Scheraga, A molecular switch for biochemical logic gates:
Conformational
studies, Biosens. Bioelectron. 1997, 12, 85-95.
7. R. Unger, J. Moult,
Towards computing with proteins, Proteins 2006, 63,
53-64.
8. M.N.
Stojanovic, D. Stefanovic, T. LaBean, H. Yan, Computing with nucleic
acids, in:
Bioelectronics: From Theory to
Applications, I. Willner, E. Katz (Eds.), Wiley-VCH, Weinheim, 2005, pp. 427-455.
9. A.
Saghatelian, N.H. Volcker, K.M. Guckian, V.S.Y. Lin, M.R. Ghadiri,
DNA-based
photonic logic gates: AND, NAND, and INHIBIT, J. Am. Chem.
Soc. 2003,
125, 346-347.
10.
G. Ashkenasy, M.R. Ghadiri, Boolean logic
functions of a synthetic peptide network,
J. Am. Chem. Soc. 2004, 126,
11140-11141.
11. M.N. Win, C.D. Smolke,
Higher-order cellular information
processing with synthetic RNA devices, Science
2008, 322, 456-460.
12. M.L. Simpson,
G.S. Sayler, J.T. Fleming, B. Applegate, Whole-cell biocomputing, Trends Biotechnol. 2001, 19,
317-323.
13. J.D. Gunton, R. Toral, C. Mirasso, M.E.
Gracheva, Invited review: The role of noise in some physical and
biological
systems, chapter in the book Recent Research Developments in Applied
Physics, Recent Res. Developm. Appl. Phys. 2003,
6, 497-514.
14.
R. Baron, O. Lioubashevski, E. Katz, T.
Niazov, I. Willner, Elementary
arithmetic operations by enzymes: A model for metabolic pathway based
computing, Angew. Chem.
Int. Ed. 2006, 45, 1572-1576.
15.
E. Katz, V. Privman, Enzyme-based logic
systems for information processing, Chem.
Soc. Rev. 2010, 39, 1835-1857.
16.
T. Niazov, R. Baron, E. Katz, O.
Lioubashevski, I. Willner, Concatenated logic gates using four coupled
biocatalysts operating in series, Proc.
Natl. Acad. USA. 2006,
103, 17160-17163.
17. G. Strack, M.
Ornatska, M. Pita, E. Katz, Biocomputing security system: Concatenated
enzyme-based logic gates operating as a biomolecular keypad lock, J. Am. Chem. Soc. 2008, 130,
4234-4235.
18.
E. Katz, V. Privman, J. Wang, Towards
Biosensing Strategies Based on Biochemical Logic Systems, in: Proc. Conf. ICQNM 2010, IEEE Comp. Soc.
Conf. Publ. Serv., Los Alamitos, California, 2010, pp. 1-9.
19. K.M.
Manesh, J. Halámek, M. Pita,
J. Zhou, T.K. Tam,
P. Santhosh, M.-C. Chuang,
J.R. Windmiller, D. Abidin, E. Katz, J. Wang, Enzyme
logic gates
for the digital analysis of physiological level upon injury, Biosens. Bioelectron. 2009, 24,
3569-3574.
20.
M. Pita, J. Zhou,
K.M. Manesh,
J. Halámek, E. Katz, J. Wang, Enzyme logic gates for assessing
physiological
conditions during an injury: Towards digital sensors and actuators, Sens. Actuat. B 2009, 139, 631-636.
21.
V. Privman, M.A. Arugula, J. Halámek, M.
Pita, E. Katz, Network Analysis of
Biochemical Logic for Noise Reduction and Stability: A System of Three
Coupled
Enzymatic AND Gates, J. Phys.
Chem. B 2009, 113,
5301-5310.
22.
T.K. Tam, J. Zhou, M. Pita, M. Ornatska, S.
Minko, E. Katz, Biochemically
controlled bioelectrocatalytic interface,
J. Am. Chem. Soc. 2008, 130, 10888-10889.
23.
J. Zhou, T.K. Tam, M. Pita, M.
Ornatska, S. Minko, E. Katz, Bioelectrocatylic system coupled with
enzyme-based
biocomputing ensembles performing Boolean logic operations: Approaching
“smart”
physiologically controlled biointerfaces, ACS
Appl. Mater. Interfaces, 2009, 1,
144-149.
24.
M. Privman, T.K. Tam,
M. Pita, E.
Katz, Switchable
electrode controlled by enzyme logic network system: Approaching
physiologically regulated bioelectronics, J.
Am. Chem. Soc. 2009, 131, 1314-1321.
25.
M. Krämer, M. Pita, J. Zhou, M. Ornatska, A. Poghossian,
M. J. Schöning, E. Katz, Coupling
of biocomputing systems with electronic chips: Electronic interface for
transduction of biochemical information,
J. Phys. Chem. B 2009, 113,
2573-2579.
26.
U. Alon, An
Introduction to Systems Biology. Design
Principles of Biological Circuits, Chapman
& Hall/CRC Press, Boca Raton, FL, 2007.
27.
D.B. Strukov, G.S. Snider, D.R. Stewart, R.S.
Williams, The missing memristor found, Nature
2008, 453, 80-83; J.J. Yang, M.D.
Pickett, X. Li, D.A.A. Ohlberg, D.R.
Stewart, R.S. Williams, Memristive switching mechanism for
metal/oxide/metal
nanodevices, Nature Nanotechnology 2008,
3, 429-433; L.N. Cooper, Memories and memory: a
physicist’s
approach to the brain, Int. J. Mod. Phys.
A 2000, 15, 4069-4082; T.
Munakata, Fundamentals of the new artificial
intelligence: Neural, evolutionary, fuzzy and more, 2nd Ed.,
Springer, 2008; M. Di Ventra, Y.V.
Pershin, L.O. Chua, Circuit elements with memory: memristors,
memcapacitors and
meminductors, Proceedings of the IEEE
2009, 97, 1717-1724; Y.V. Pershin, S.
La Fontaine, M. Di Ventra,
Memristive model of amoeba's learning, Phys.
Rev. E 2009, 80, Article 021926;
Y.V. Pershin, M. Di
Ventra, Spin memristive systems, Phys.
Rev. B 2008, 78, Article 113309.
28.
A.P. de Silva, S. Uchiyama, T.P. Vance, B.
Wannalerse, A supramolecular chemistry basis for molecular logic and
computation, Coord. Chem. Rev. 2007, 251, 1623-1632; A.P. de Silva, S. Uchiyama, Molecular
logic and
computing, Nature Nanotech. 2007, 2, 399-410; K. Szacilowski, Digital information
processing in
molecular systems, Chem. Rev. 2008, 108, 3481-3548.
29.
P. Dittrich, Chemical computing, Lect. Notes
Computer Sci. 2005, 3566, 19-32.
30.
A.N. Shipway, E. Katz, I. Willner, Molecular
memory and processing devices in solution and on surfaces, in: Structure and Bonding, volume title “Molecular
Machines and Motors,” J.-P.
Sauvage (Ed.), Springer-Verlag, Berlin, 2001,
Vol. 99, pp. 237-281.
31.
M. Suresh, A. Ghosh, A. Das, Half-subtractor
operation in pH responsive N-heterocyclic
amines, Tetrahedron Lett. 2007, 48, 8205-8208.
32.
S. Iwata, K. Tanaka, A novel cation “AND” anion recognition
host having pyrido[1',2': 1,2]-imidazo[4,5-b]pyrazine
as the fluorophore, J. Chem. Soc., Chem. Commun. 1995,
1491-1492.
33.
S.H. Lee, J.Y. Kim, S.K. Kim, J.H. Leed, J.S.
Kim, Pyrene-appended calix[4]crowned logic gates involving normal and
reverse
PET: NOR, XNOR and INHIBIT, Tetrahedron 2004,
60, 5171-5176.
34.
D.C. Magri, G.J. Brown, G.D. McClean, A.P. de
Silva, Communicating chemical congregation: A molecular AND logic gate
with
three chemical inputs as a ‘Lab-on-a-Molecule’ prototype, J.
Am. Chem. Soc. 2006, 128,
4950-4951.
35.
F.M. Raymo, S. Giordani,
All-optical processing with molecular switches, Proc. Natl. Acad.
USA
2002, 99, 4941-4944.
36. G. Nishimura, K. Ishizumi, Y.
Shiraishi, T. Hirai, A
triethylenetetramine with anthracene and benzophenone as a fluorescent
molecular logic gate with Either-Or switchable dual logic functions, J. Phys. Chem. B 2006, 110,
21596-21602.
37.
Y. Shiraishi, Y. Tokitoh, T. Hirai, A
fluorescent molecular logic gate with multiply-configurable dual
outputs, Chem. Commun. 2005,
5316-5318.
38.
R. Baron, A. Onopriyenko, E. Katz, O.
Lioubashevski, I. Willner, S. Wang, H. Tian, An
electrochemical/photochemical information processing system using a
monolayer-functionalized electrode,,Chem.
Commun. 2006,
2147-2149.
39. M. Biancardo, C. Bignozzi, H. Doyle, G.
Redmond, A potential and ion switched molecular photonic logic gate, Chem. Commun. 2005,
3918-3920.
40.
J.H. Qian, Y.F. Xu, X.H. Qian, S.Y. Zhang, Molecular logic
operations based on surfactant nanoaggregates, ChemPhysChem
2008, 9, 1891-1898.
41.
A.P. de Silva, Molecular computing – a layer
of logic, Nature 2008, 454,
417-418.
42.
S. Nitahara, N. Terasaki, T. Akiyama, S.
Yamada, Molecular logic device using mixed self-assembled monolayers, Thin Solid Films 2006, 499,
354-358.
43.
T. Gupta, M.E. van der Boom, Redox-active
monolayers as a versatile platform for integrating Boolean logic gates, Angew. Chem. Int. Ed. 2008, 47, 5322-5326.
44. A. Credi,
V. Balzani, S.J. Langford, J.F. Stoddart, Logic operations at the
molecular
level. An XOR gate based on a molecular machine, J.
Am. Chem. Soc. 1997, 119, 2679-2681.
45.
A.P. de Silva, H.Q.N. Gunaratne, C.P. McCoy,
A molecular photoionic AND gate based on fluorescent signalling, Nature 1993, 364,
42-44; A.P. de
Silva, H.Q.N. Gunaratne, C.P. McCoy, Molecular photoionic AND logic
gates with
bright fluorescence and "Off-On" digital action, J. Am.
Chem. Soc. 1997, 119, 7891-7892.
46.
A.P. de Silva, H.Q.N. Gunaratne, G.E.M.
Maguire, 'Off-on' fluorescent sensors for physiological
levels of
magnesium ions based on photoinduced electron transfer (PET),
which
also behave as photoionic OR logic gates, J.
Chem. Soc., Chem. Commun. 1994,
1213-1214.
47.
A.P. de Silva, N.D. McClenaghan,
Simultaneously multiply-configurable or superposed molecular logic
systems
composed of ICT (Internal Charge Transfer) chromophores and
fluorophores
integrated with one or two ion receptors, Chem.
Eur. J. 2002, 8, 4935-4945.
48.
A.P. de Silva, I.M. Dixon, H.Q.N. Gunaratne,
T. Gunnlaugsson, P.R.S. Maxwell, T.E. Rice, Integration of logic
functions and
sequential operation of gates at the molecular-scale, J.
Am. Chem. Soc. 1999, 121,
1393-1394.
49.
S.D. Straight, P.A.
Liddell, Y.
Terazono, T.A. Moore, A.L.
Moore, D. Gust, All-photonic molecular
XOR and NOR logic gates based on
photochemical control of fluorescence in a
fulgimide-porphyrin-dithienylethene
triad, Adv. Funct. Mater. 2007, 17, 777–785.
50.
Z. Wang, G. Zheng, P. Lu,
9-(Cycloheptatrienylidene)-fluorene derivative: Remarkable ratiometric
pH
sensor and computing switch with NOR logic gate, Org. Lett.
2005, 7, 3669-3672.
51.
H.T. Baytekin, E.U. Akkaya, A molecular NAND
gate based on Watson-Crick base-pairing, Org.
Lett. 2000, 2, 1725-1727.
52.
G. Zong, L. Xiana, G. Lua, l-Arginine bearing
an anthrylmethyl group: fluorescent molecular NAND logic gate with H+
and ATP
as inputs, Tetrahedron Lett. 2007, 48, 3891-3894.
53.
T. Gunnlaugsson, D.A. MacDónaill, D. Parker,
Lanthanide macrocyclic quinolyl conjugates as luminescent
molecular-level
devices, J. Am. Chem. Soc. 2001, 123, 12866-12876; T. Gunnlaugsson, D.A.
MacDónaill, D. Parker, Luminescent molecular logic
gates: the
two-input inhibit (INH) function, Chem.
Commun. 2000,
93-94
54.
M. de Sousa, B. de Castro, S. Abad, M.A.
Miranda, U. Pischel, A
molecular tool kit for the variable design of logic operations (NOR,
INH,
EnNOR), Chem. Commun.
2006, 2051-2053.
55.
L. Li, M.-X. Yu, F.Y. Li, T. Yi, C.H. Huang,
INHIBIT logic gate based on spiropyran sensitized semiconductor
electrode, Colloids Surf. A 2007, 304, 49-53.
56. V. Luxami, S. Kumar, Molecular half-subtractor
based on
3,3'-bis(1H-benzimidazolyl-2-yl)[1,1']binaphthalenyl-2,2'-diol, New J. Chem. 2008, 32,
2074-2079.
57. J.H. Qian, X.H. Qian, Y.F. Xu,
S.Y. Zhang,
Multiple
molecular logic functions and molecular calculations facilitated by
surfactants’ versatility, Chem.
Commun. 2008, 4141-4143.
58.
E. Pérez-Inestrosa, J.-M.
Montenegro, D. Collado, R. Suau,
J. Casado, Molecules with
multiple light-emissive electronic excited states as a strategy toward
molecular reversible logic gates, J. Phys.
Chem. C 2007, 111,
6904-6909.
59. W. Sun, C.H. Xu, Z. Zhu, C.J.
Fang, C.H. Yan,
Chemical-driven reconfigurable arithmetic functionalities within a
fluorescent
tetrathiafulvalene derivative, J. Phys.
Chem. C 2008, 112, 16973-16983;
Z.-X. Li, L.-Y. Liao,
W. Sun, C.-H. Xu, C. Zhang, C.-J. Fang, C.-H. Yan, Reconfigurable
cascade
circuit in a photo- and chemical-switchable fluorescent diarylethene
derivative, J. Phys. Chem. C 2008, 112, 5190-5196.
60.
A. Coskun, E. Deniz, E.U. Akkaya, Effective
PET and ICT switching of boradiazaindacene emission: A unimolecular,
emission-mode, molecular half-subtractor with reconfigurable logic
gates, Org. Lett. 2005, 7,
5187-5189.
61.
D. Jimenez, R. Martinez-Manez, F. Sancenon,
J.V. Ros-Lis, J. Soto, A. Benito, E. Garcia-Breijo, Multi-channel
receptors and
their relation to guest chemosensing and reconfigurable molecular logic
gates, Eur. J. Inorg. Chem. 2005,
2393-2403.
62. W. Sun, Y.-R. Zheng, C.-H. Xu, C.-J.
Fang,
C.-H. Yan, Fluorescence-based reconfigurable and resettable molecular
arithmetic mode, J. Phys. Chem. C 2007,
111, 11706-11711.
63. Y. Zhou, H. Wu, L. Qu, D. Zhang, D. Zhu,
A
new redox-resettable molecule-based half-adder with tetrathiafulvalene,
J.
Phys. Chem. B 2006, 110,
15676-15679.
64.
U. Pischel, B. Heller, Molecular logic devices
(half-subtractor, comparator, complementary output circuit) by
controlling
photoinduced charge transfer processes, New
J. Chem. 2008, 32, 395-400.
65. J.
Andreasson, S.D. Straight, S.
Bandyopadhyay, R.H. Mitchell, T.A. Moore, A.L. Moore, D. Gust, A
molecule-based
1:2 digital demultiplexer, J. Phys. Chem.
C 2007, 111, 14274-14278; M.
Amelia, M. Baroncini, A. Credi, A simple unimolecular
multiplexer/demultiplexer, Angew. Chem. Int. Ed. 2008, 47, 6240-6243; E. Perez-Inestrosa,
J.M. Montenegro, D. Collado, R.
Suau, A molecular 1 : 2
demultiplexer, Chem.
Commun. 2008, 1085-1087.
66. J. Andreasson, S.D. Straight,
T.A. Moore, A.L. Moore, D.
Gust, Molecular all-photonic encoder−decoder, J. Am. Chem.
Soc. 2008, 130, 11122-11128.
67.
D. Margulies, C.E. Felder, G. Melman, A.
Shanzer, A molecular keypad lock: A photochemical device capable of authorizing password entries, J. Am. Chem.
Soc. 2007, 129, 347-354; M. Suresh,
A. Ghosh, A.
Das, A simple
chemosensor for Hg2+ and Cu2+ that works as a
molecular
keypad lock, Chem.
Commun. 2008, 3906-3908.
68. R. Baron, A. Onopriyenko, E. Katz, O.
Lioubashevski, I. Willner, S. Wang, H. Tian, An
electrochemical/photochemical
information processing system using a monolayer-functionalized
electrode, Chem. Commun. 2006, 2147-2149.
69. E. Katz, I. Willner, A quinone-functionalized
electrode in
conjunction with hydrophobic magnetic nanoparticles acts as a
“Write–Read–Erase” information storage system, Chem.
Commun. 2005, 5641-5643.
70. E. Katz, I. Willner, A bis-quinone-functionalized
gold-electrode subjected to hydrophobic magnetic nanoparticles acts as
a
three-state "Write-Read-Erase" information storage system, Electrochem.
Commun. 2006, 8, 879-882.
71. F. Galindo, J.C. Lima, S.V. Luis, A.J.
Parola, F. Pina, Write-Read-Erase molecular-switching system trapped in
a
polymer hydrogel matrix, Adv. Funct.
Mater. 2005, 15, 541-545.
72. A. Bandyopadhyay, A.J. Pal,
Memory-switching
phenomenon in acceptor-rich organic molecules: Impedance spectroscopic
studies,
J. Phys. Chem. B 2005, 109,
6084-6088.
73. F. Pina, J.C. Lima, A.J. Parola, C.A.M.
Afonso, Thermal
and photochemical properties of 4',7-dihydroxyflavylium in water-ionic
liquid
biphasic systems: A Write-Read-Erase molecular switch, Angew. Chem. Int. Ed. 2004,
43, 1525-1527.
74. G. Will, J.S.S.N. Rao, D. Fitzmaurice, Heterosupramolecular
optical
write–read–erase device, J. Mater.
Chem. 1999, 9, 2297-2299.
75. J. Hiller, M.F. Rubner,
Reversible
molecular memory and pH-switchable swelling transitions in
polyelectrolyte
multilayers, Macromolecules 2003, 36, 4078-4083.
76. F. Pina, A. Roque, M.J. Melo, I.
Maestri, L.
Belladelli, V. Balzani, Multistate/multifunctional molecular-level
systems:
light and pH switching between the various forms of a synthetic
flavylium salt,
Chem. Eur. J. 1998, 4,
1184-1191.
77.
U. Pischel, Chemical approaches to
molecular logic elements for
addition and subtraction, Angew.
Chem. Int. Ed. 2007, 46,
4026-4040.
78.
G.J. Brown, A.P. de Silva, S. Pagliari, Molecules that add up,
Chem. Commun. 2002, 2461-2463.
79.
D.-H. Qu, Q.-C. Wang, H. Tian, A half adder based on a
photochemically driven [2]rotaxane, Angew.
Chem. Int. Ed. 2005,
44, 5296-5299.
80.
J. Andréasson, S.D. Straight, G. Kodis, C.-D.
Park, M. Hambourger, M. Gervaldo, B. Albinsson, T. A. Moore, A.L.
Moore, D.
Gust, All-photonic molecular half-adder, J.
Am. Chem. Soc. 2006, 128,
16259-16265; J. Andréasson, G.
Kodis, Y. Terazono, P.A. Liddell, S. Bandyopadhyay, R.H. Mitchell, T.A.
Moore,
A.L. Moore, D. Gust, Molecule-based photonically switched half-adder, J. Am. Chem. Soc. 2004, 126,
15926-15927.
81.
M.V. Lopez, M.E. Vazquez, C. Gomez-Reino, R.
Pedrido, M.R. Bermejo, A
metallo-supramolecular approach to a half-subtractor, New J. Chem. 2008, 32,
1473-1477.
82.
D. Margulies, C.E. Felder, G. Melman, A.
Shanzer, A molecular keypad lock: A photochemical device capable of
authorizing
password entries, J. Am. Chem. Soc. 2006,
128, 4865-4871.
83.
O. Kuznetz, H. Salman, N. Shakkour, Y.
Eichen, S. Speiser, A novel all optical molecular scale full adder, Chem. Phys. Lett. 2008, 451,
63-67.
84.
Y. Liu, W. Jiang, H.-Y. Zhang,
C.-J. Li, A multifunctional arithmetical processor model integrated
inside a
single molecule, J.
Phys. Chem. B 2006, 110, 14231-14235.
85.
X. Guo, D. Zhang, G. Zhang, D.
Zhu, Monomolecular logic: “Half-Adder” based on
multistate/multifunctional
photochromic spiropyrans, J.
Phys. Chem. B 2004, 108, 11942-11945.
86. F.
Pina, M.J. Melo, M. Maestri, P. Passaniti, V. Balzani, Artificial
chemical
systems capable of mimicking some elementary properties of neurons, J.
Am.
Chem. Soc. 2000, 122,
4496-4498.
87.
R. Stadler, S. Ami, C. Joachim, M. Forshaw, Integrating logic functions
inside a
single molecule, Nanotechnology
2004, 15, S115-S121; F.M. Raymo, S.
Giordani, Signal processing at the
molecular level, J. Am. Chem. Soc. 2001,
123, 4651-4652.
88. A.P. De Silva, Y. Leydet, C. Lincheneau,
N.D.
McClenaghan, Chemical approaches to nanometer-scale logic gates, J. Phys. Cond. Matter 2006, 18, S1847-S1872.
89. A. Adamatzky, Computing
with waves in chemical media: Massively parallel reaction-diffusion
processors,
IEICE Trans. Electronics 2004, E87C, 1748-1756.
90. A.H. Flood, R.J.A. Ramirez, W.Q. Deng,
R.P.
Muller, W.A. Goddard, J.F. Stoddart, Meccano on the nanoscale - A
blueprint for
making some of the world’s tiniest machines, Austr. J.
Chem. 2004, 57,
301-322.
91. J. Xu,
G.J. Tan, A
review on DNA computing models, J.
Comput. Theor. Nanosci. 2007, 4,
1219-1230.
92. M.
Kahan, B. Gil, R.
Adar, E. Shapiro, Towards molecular computers that operate in a
biological
environment, Physica D 2008, 237, 1165-1172.
93.
X.-G. Shao, H.-Y. Jiang, W.-S. Cai, Advances in biomolecular
computing, Prog. Chem. 2002,
14, 37-46.
94.
D. Melnikov, G. Strack, M. Pita, V. Privman,
E. Katz, Analog noise reduction in enzymatic logic gates, J.
Phys. Chem. B 2009, 113,
10472-10479.
95.
L. Fedichkin, E. Katz, V. Privman, Error
correction and digitalization concepts
in biochemical computing, J. Comput. Theor. Nanosci 2008,
5, 36-43.
