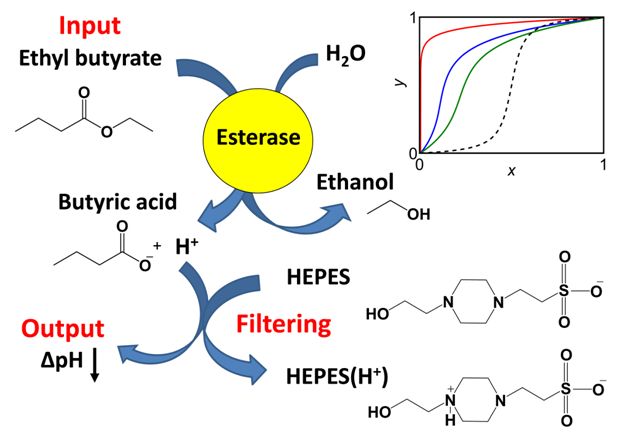
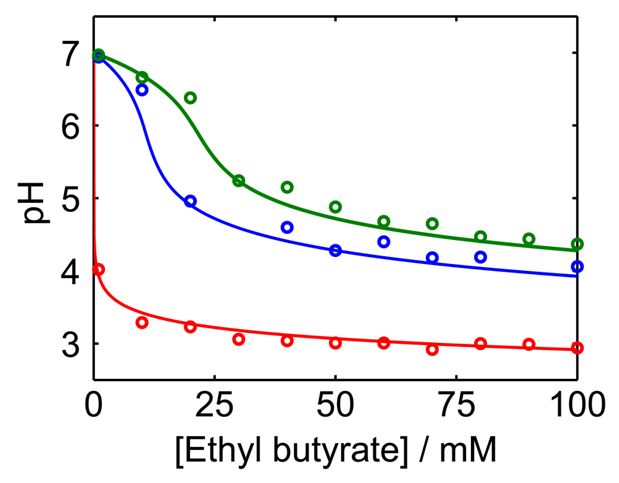
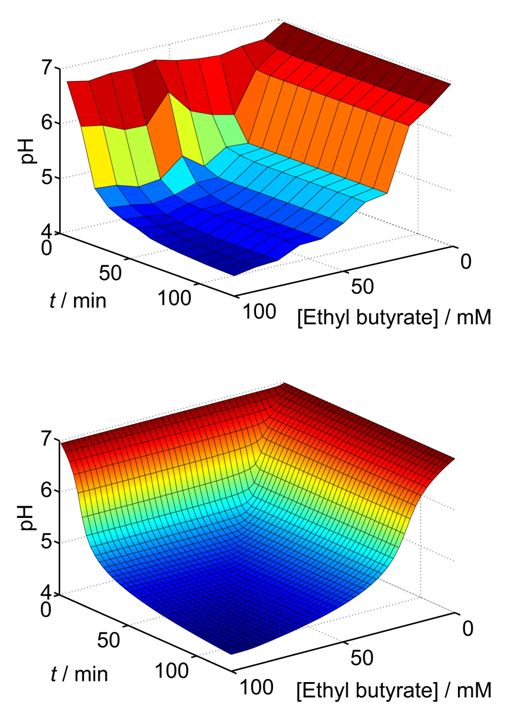
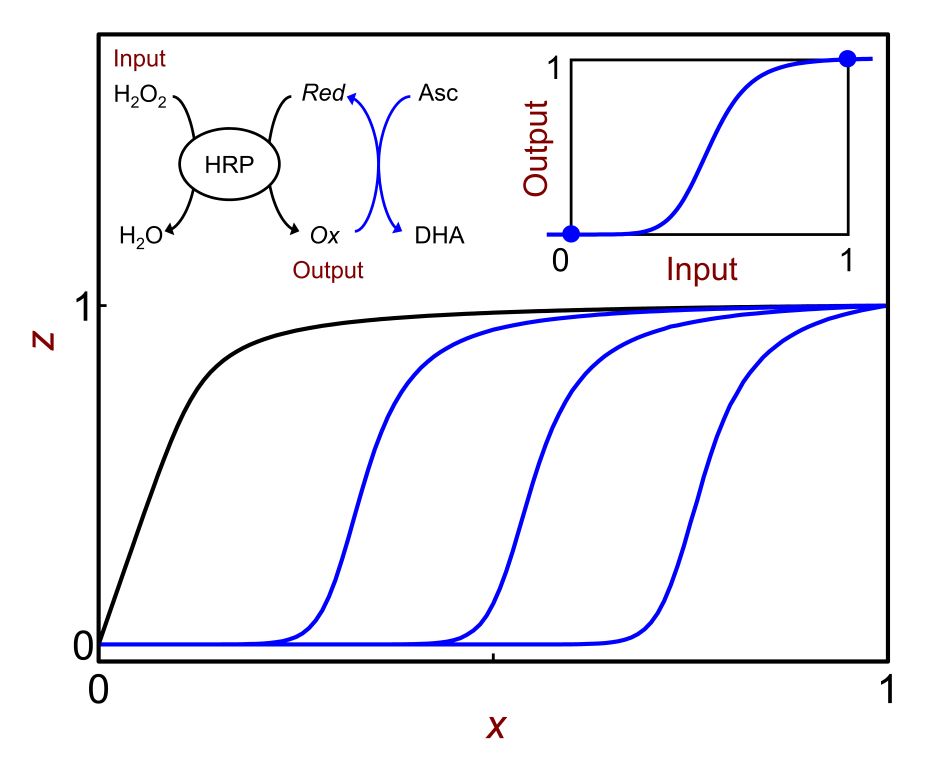
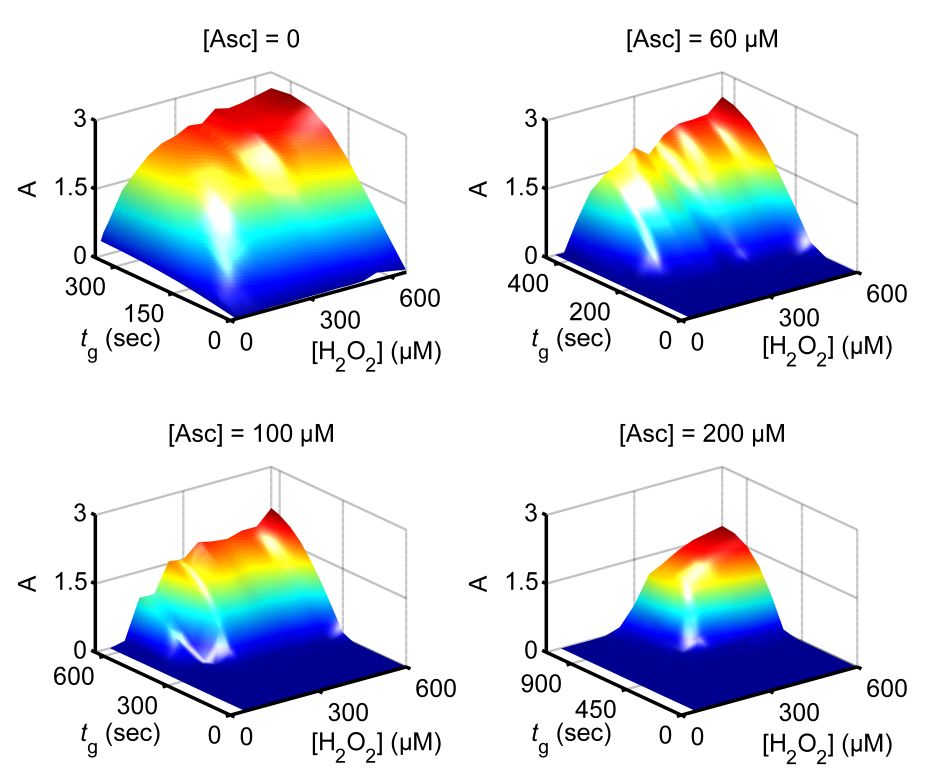
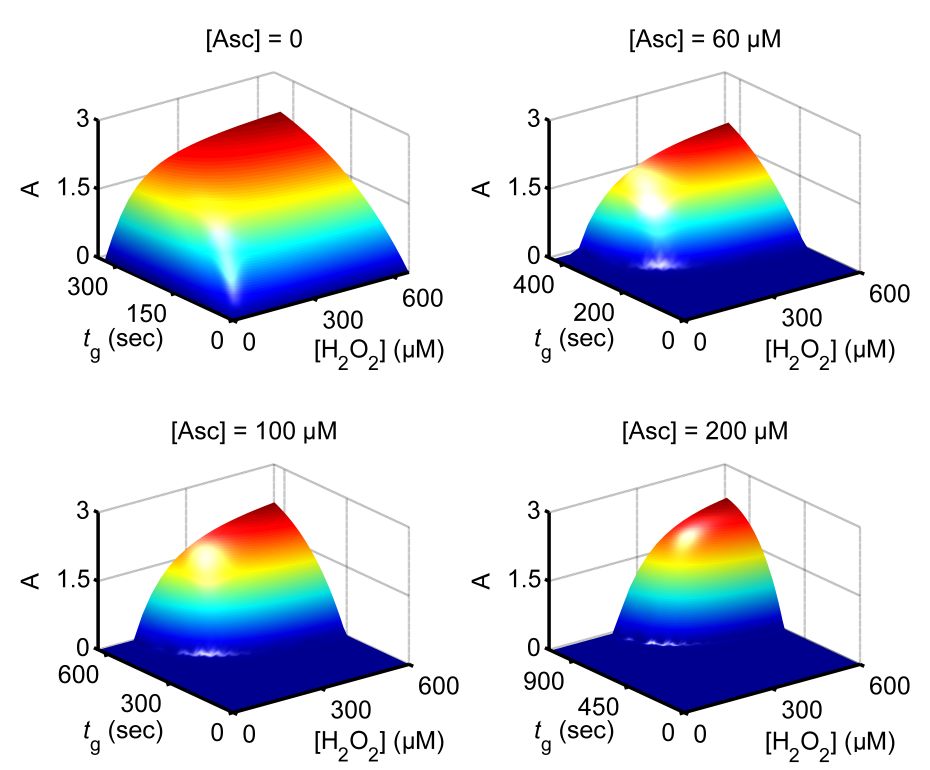
This research project is conducted in a close collaboration with Prof. Vladimir Privman
PI: Evgeny Katz, Co-PI: Vladimir Privman
Title: "Biochemical Computing: Experimental and Theoretical Development of Error Correction and Digitalization Concepts"
Agency: National Science Foundation (NSF)
Award No: CCF-0726698
Time Period: 09/15/07 - 08/31/10
Title: "SHF: Small: Experimental and Theoretical Development of Error Correction and Digitalization Concepts for Multi-Enzyme Biomolecular Computing Networks"
Agency: National Science Foundation (NSF)
Award No: CCF-1015983
Time Period: 09/01/10 - 08/31/13
The need for a “toolbox” of non-Boolean network elements: Why are non-Boolean network elements, hopefully those with modular properties (that can be reused in various network designs), needed? There are actually several different aspects of networking that require such functionalities: Digital error correction for larger networks (which for biochemical logic sometimes means networks involving as few as order 10 information processing steps), is based on redundancy and therefore requires at least the elements for splitting the signal, for signal amplification and intensity-balancing, which, for biochemical logic is easier accomplished by utilizing signal attenuation. Filtering is obviously useful for control of analog errors, by resetting the signals to their reference digital values. Signal resetting is more generally a useful function if we were to attempt to “clock” the biocomputing processes, in future information processing designs. Finally, converters between various types of chemical “signals” will also be useful for networking. Among the more risky research directions that we hope to explore, are bio-inspired network elements involving time-delay and memory.